A New Genome Editing Technique
The CRISPR/Cas9 system is currently the technique in genome editing. Thanks to recent developments in the CRISPR genome editing system, we are able to alter DNA with unprecedented precision and accuracy. Ultimately, this revolutionary genome editing technique allows us to modify any region of the genome of any species—without harming other genes. But more than that, we are able to edit these genes at just a fraction of the cost of previous methods.
It has utterly revolutionized gene editing.
However, a team of researchers has just developed a new approach that just might prove to be as efficient and effective as the current standard.
In a study published in Nature Biotechnology, researchers from Chunyu Han’s lab have developed a novel genome editing technique basically just like CRISPR.
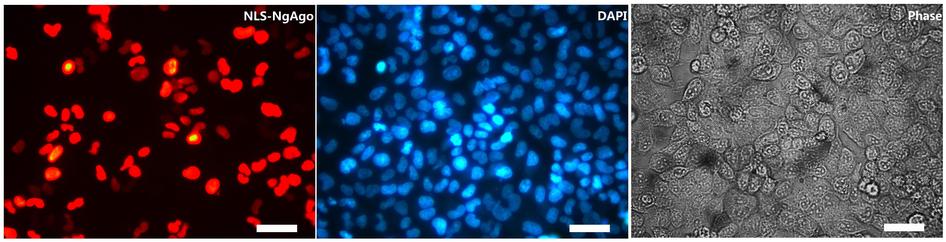
According to the report, the method is based on the Natronobacterium gregoryi Argonaute (NgAgo), a DNA-guided endonuclease that’s similar to Cas9, the endonuclease of CRISPR. These are a specific type of protein, otherwise known as restriction enzymes, that are responsible for cutting DNA at specific locations.
Remarkably, the study shows that NgAgo is suitable for genome editing in human cells.
Beating CRISPR?
The team asserts that their study has a number of key differences between NgAgo and Cas9, where the former is at the advantage. They claim that that the method appears to have a low tolerance to guide-target mismatches, leading to a high efficiency in editing.
One of these is that NgAgo does not need to be followed by a protospacer-adjacent motif (PAM), a DNA sequence seen in Cas9. Notably, Cas9 will not successfully bind or cleave a target DNA sequence if this is not followed by the PAM (as it is responsible for the protein to differentiate CRISPR DNA from target DNA).
Another benefit is that the loading temperature of NgAgo is found to be at 55°C and not at Cas9’s 37°C. This shows that this could be another option for genome editing at conditions such as this. Also, at this temperature, the method follows a “one-guide-faithful” rule, that is, NgAgo cannot swap target DNA with other free DNA, minimizing off-target effects.
Though there has yet to be an extensive side-by-side comparison of the two enzymes, NgAgo and Cas9 appear to have similar efficiencies. The authors report that their tests with 47 guides targeting 8 human genes, the results showed a 21% to 41% efficiency.